SegmentationImage¶
- class photutils.segmentation.SegmentationImage(data)[source]¶
Bases:
objectClass for a segmentation image.
- Parameters:
- data2D int
ndarray A 2D segmentation array where source regions are labeled by different positive integer values. A value of zero is reserved for the background. The segmentation image must have integer type.
- data2D int
Notes
The
SegmentationImageinstance may be sliced, but note that the slicedSegmentationImagedata array will be a view into the originalSegmentationImagearray (this is the same behavior asnumpy.ndarray). Explicitly use theSegmentationImage.copy()method to create a copy of the slicedSegmentationImage.Attributes Summary
A 1D array of areas (in pixel**2) of the non-zero labeled regions.
The area (in pixel**2) of the background (label=0) region.
A list of
BoundingBoxof the minimal bounding boxes containing the labeled regions.A matplotlib colormap consisting of (random) muted colors.
The segmentation array.
A
MaskedArrayversion of the segmentation array where the background (label = 0) has been masked.Boolean value indicating whether or not the non-zero labels in the segmentation array are consecutive and start from 1.
The sorted non-zero labels in the segmentation array.
The maximum label in the segmentation array.
A 1D
ndarrayof the sorted non-zero labels that are missing in the consecutive sequence from one to the maximum label number.The number of non-zero labels in the segmentation array.
A list of Shapely polygons representing each source segment.
A list of
Segmentobjects.The shape of the segmentation array.
A list of tuples, where each tuple contains two slices representing the minimal box that contains the labeled region.
Methods Summary
check_label(label)Check that the input label is a valid label number within the segmentation array.
check_labels(labels)Check that the input label(s) are valid label numbers within the segmentation array.
copy()Return a deep copy of this class instance.
get_area(label)The area (in pixel**2) of the region for the input label.
get_areas(labels)The areas (in pixel**2) of the regions for the input labels.
get_index(label)Find the index of the input
label.get_indices(labels)Find the indices of the input
labels.imshow([ax, figsize, dpi, cmap, alpha])Display the segmentation image in a matplotlib
Axesinstance.imshow_map([ax, figsize, dpi, cmap, alpha, ...])Display the segmentation image in a matplotlib
Axesinstance with a colorbar.keep_label(label[, relabel])Keep only the specified label.
keep_labels(labels[, relabel])Keep only the specified labels.
make_cmap([background_color, seed])Define a matplotlib colormap consisting of (random) muted colors.
make_source_mask(*[, size, footprint])Make a source mask from the segmentation image.
plot_patches(*[, ax, origin, scale, labels])Plot the
Polygonobjects for the source segments on a matplotlibAxesinstance.reassign_label(label, new_label[, relabel])Reassign a label number to a new number.
reassign_labels(labels, new_label[, relabel])Reassign one or more label numbers.
relabel_consecutive([start_label])Reassign the label numbers consecutively starting from a given label number.
remove_border_labels(border_width[, ...])Remove labeled segments near the array border.
remove_label(label[, relabel])Remove the label number.
remove_labels(labels[, relabel])Remove one or more labels.
remove_masked_labels(mask[, ...])Remove labeled segments located within a masked region.
reset_cmap([seed])Reset the colormap (
cmapattribute) to a new random colormap.to_patches(*[, origin, scale])Return a list of
Polygonobjects representing each source segment.Attributes Documentation
- areas¶
A 1D array of areas (in pixel**2) of the non-zero labeled regions.
The
ndarraystarts with the non-zero label. The returned array has a length equal to the number of labels and matches the order of thelabelsattribute.
- background_area¶
The area (in pixel**2) of the background (label=0) region.
- bbox¶
A list of
BoundingBoxof the minimal bounding boxes containing the labeled regions.
- cmap¶
A matplotlib colormap consisting of (random) muted colors.
This is useful for plotting the segmentation array.
- data¶
The segmentation array.
- data_ma¶
A
MaskedArrayversion of the segmentation array where the background (label = 0) has been masked.
- is_consecutive¶
Boolean value indicating whether or not the non-zero labels in the segmentation array are consecutive and start from 1.
- labels¶
The sorted non-zero labels in the segmentation array.
- max_label¶
The maximum label in the segmentation array.
- missing_labels¶
A 1D
ndarrayof the sorted non-zero labels that are missing in the consecutive sequence from one to the maximum label number.
- nlabels¶
The number of non-zero labels in the segmentation array.
- segments¶
A list of
Segmentobjects.The list starts with the non-zero label. The returned list has a length equal to the number of labels and matches the order of the
labelsattribute.
- shape¶
The shape of the segmentation array.
- slices¶
A list of tuples, where each tuple contains two slices representing the minimal box that contains the labeled region.
The list starts with the non-zero label. The returned list has a length equal to the number of labels and matches the order of the
labelsattribute.
Methods Documentation
- check_label(label)[source]¶
Check that the input label is a valid label number within the segmentation array.
- Parameters:
- labelint
The label number to check.
- Raises:
- ValueError
If the input
labelis invalid.
- check_labels(labels)[source]¶
Check that the input label(s) are valid label numbers within the segmentation array.
- Parameters:
- labelsint, 1D array_like (int)
The label(s) to check.
- Raises:
- ValueError
If any input
labelsare invalid.
- get_area(label)[source]¶
The area (in pixel**2) of the region for the input label.
- Parameters:
- labelint
The label whose area to return. Label must be non-zero.
- Returns:
- areafloat
The area of the labeled region.
- get_areas(labels)[source]¶
The areas (in pixel**2) of the regions for the input labels.
- Parameters:
- labelsint, 1D array_like (int)
The label(s) for which to return areas. Label must be non-zero.
- Returns:
- areas
ndarray The areas of the labeled regions.
- areas
- get_index(label)[source]¶
Find the index of the input
label.- Parameters:
- labelint
The label number to find.
- Returns:
- indexint
The array index.
- Raises:
- ValueError
If
labelis invalid.
- get_indices(labels)[source]¶
Find the indices of the input
labels.- Parameters:
- labelsint, array_like (1D, int)
The label numbers(s) to find.
- Returns:
- indicesint
ndarray An integer array of indices with the same shape as
labels. Iflabelsis a scalar, then the returned index will also be a scalar.
- indicesint
- Raises:
- ValueError
If any input
labelsare invalid.
- imshow(ax=None, figsize=None, dpi=None, cmap=None, alpha=None)[source]¶
Display the segmentation image in a matplotlib
Axesinstance.The segmentation image will be displayed with “nearest” interpolation and with the origin set to “lower”.
- Parameters:
- ax
matplotlib.axes.AxesorNone, optional The matplotlib axes on which to plot. If
None, then a newAxesinstance will be created.- figsize2-tuple of floats or
None, optional The figure dimension (width, height) in inches when creating a new Axes. This keyword is ignored if
axesis input.- dpifloat or
None, optional The figure dots per inch when creating a new Axes. This keyword is ignored if
axesis input.- cmap
matplotlib.colors.Colormap, str, orNone, optional The
Colormapinstance or a registered matplotlib colormap name used to map scalar data to colors. IfNone, then the colormap defined by thecmapattribute will be used.- alphafloat, array_like, or
None, optional The alpha blending value, between 0 (transparent) and 1 (opaque). If alpha is an array, the alpha blending values are applied pixel by pixel, and alpha must have the same shape as the segmentation image.
- ax
- Returns:
- result
matplotlib.image.AxesImage An image attached to an
matplotlib.axes.Axes.
- result
Examples
import numpy as np from photutils.segmentation import SegmentationImage data = np.array([[1, 1, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]]) segm = SegmentationImage(data) fig, ax = plt.subplots() im = segm.imshow(ax=ax) fig.colorbar(im, ax=ax)
(
Source code,png,hires.png,pdf,svg)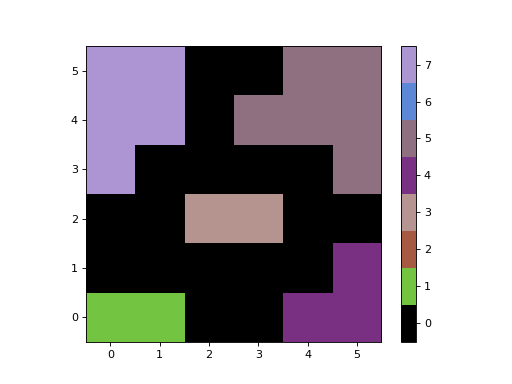
- imshow_map(ax=None, figsize=None, dpi=None, cmap=None, alpha=None, max_labels=25, cbar_labelsize=None)[source]¶
Display the segmentation image in a matplotlib
Axesinstance with a colorbar.This method is useful for displaying segmentation images that have a small number of labels (e.g., from a cutout) that are not consecutive. It maps the labels to be consecutive integers starting from 1 before plotting. The plotted image values are not the label values, but the colorbar tick labels are used to show the original labels.
The segmentation image will be displayed with “nearest” interpolation and with the origin set to “lower”.
- Parameters:
- ax
matplotlib.axes.AxesorNone, optional The matplotlib axes on which to plot. If
None, then a newAxesinstance will be created.- figsize2-tuple of floats or
None, optional The figure dimension (width, height) in inches when creating a new Axes. This keyword is ignored if
axesis input.- dpifloat or
None, optional The figure dots per inch when creating a new Axes. This keyword is ignored if
axesis input.- cmap
matplotlib.colors.Colormap, str, orNone, optional The
Colormapinstance or a registered matplotlib colormap name used to map scalar data to colors. IfNone, then the colormap defined by thecmapattribute will be used.- alphafloat, array_like, or
None, optional The alpha blending value, between 0 (transparent) and 1 (opaque). If alpha is an array, the alpha blending values are applied pixel by pixel, and alpha must have the same shape as the segmentation image.
- max_labelsint, optional
The maximum number of labels to display in the colorbar. If the number of labels is greater than
max_labels, then the colorbar will not be displayed.- cbar_labelsize
Noneor float, optional The font size of the colorbar tick labels.
- ax
- Returns:
- result
matplotlib.image.AxesImage An image attached to an
matplotlib.axes.Axes.- cbar_infotuple or
None The colorbar information as a tuple containing the
Colorbarinstance, andarrayof tick positions, and andarrayof tick labels.Noneis returned if the colorbar was not plotted.
- result
Examples
import numpy as np from photutils.segmentation import SegmentationImage data = np.array([[1, 1, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]]) data *= 1000 segm = SegmentationImage(data) fig, ax = plt.subplots() im, cbar = segm.imshow_map(ax=ax)
(
Source code,png,hires.png,pdf,svg)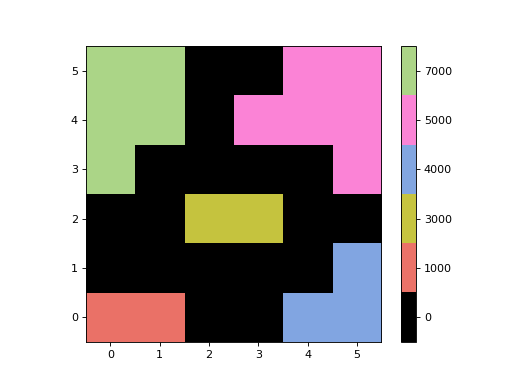
- keep_label(label, relabel=False)[source]¶
Keep only the specified label.
- Parameters:
- labelint
The label number to keep.
- relabelbool, optional
If
True, then the single segment will be assigned a label value of 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.keep_label(label=3) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.keep_label(label=3, relabel=True) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 1, 1, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]])
- keep_labels(labels, relabel=False)[source]¶
Keep only the specified labels.
- Parameters:
- labelsint, array_like (1D, int)
The label number(s) to keep.
- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.keep_labels(labels=[5, 3]) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 0, 0], [0, 0, 0, 0, 0, 5], [0, 0, 0, 5, 5, 5], [0, 0, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.keep_labels(labels=[5, 3], relabel=True) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 1, 1, 0, 0], [0, 0, 0, 0, 0, 2], [0, 0, 0, 2, 2, 2], [0, 0, 0, 0, 2, 2]])
- make_cmap(background_color='#000000ff', seed=None)[source]¶
Define a matplotlib colormap consisting of (random) muted colors.
This is useful for plotting the segmentation array.
- Parameters:
- background_colorMatplotlib color, optional
The color of the first color in the colormap. The color may be specified using any of the Matplotlib color formats. This color will be used as the background color (label = 0) when plotting the segmentation image. The default color is black with alpha=1.0 (‘#000000ff’).
- seedint, optional
A seed to initialize the
numpy.random.BitGenerator. IfNone, then fresh, unpredictable entropy will be pulled from the OS. Separate function calls with the sameseedwill generate the same colormap.
- Returns:
- cmap
matplotlib.colors.ListedColormap The matplotlib colormap with colors in RGBA format.
- cmap
- make_source_mask(*, size=None, footprint=None)[source]¶
Make a source mask from the segmentation image.
Use the
sizeorfootprintkeyword to perform binary dilation on the segmentation image mask.- Parameters:
- sizeint or tuple of int, optional
The size along each axis of the rectangular footprint used for the source dilation. If
sizeis a scalar, then a square footprint ofsizewill be used. Ifsizehas two elements, they must be in(ny, nx)order.sizeshould have odd values for each axis. To perform source dilation, eithersizeorfootprintmust be defined. If they are both defined, thenfootprintoverridessize.- footprint2D
ndarray, optional The local footprint used for the source dilation. Non-zero elements are considered
True.size=(n, m)is equivalent tofootprint=np.ones((n, m)). To perform source dilation, eithersizeorfootprintmust be defined. If they are both defined, thenfootprintoverridessize.
- Returns:
- mask2D bool
ndarray A 2D boolean image containing the source mask.
- mask2D bool
Notes
When performing source dilation, using a square footprint will be much faster than using other shapes (e.g., a circular footprint). Source dilation also is slower for larger images and larger footprints.
Examples
>>> import numpy as np >>> from photutils.segmentation import SegmentationImage >>> from photutils.utils import circular_footprint >>> data = np.zeros((7, 7), dtype=int) >>> data[3, 3] = 1 >>> segm = SegmentationImage(data) >>> segm.data array([[0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 1, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0]])
>>> mask0 = segm.make_source_mask() >>> mask0 array([[False, False, False, False, False, False, False], [False, False, False, False, False, False, False], [False, False, False, False, False, False, False], [False, False, False, True, False, False, False], [False, False, False, False, False, False, False], [False, False, False, False, False, False, False], [False, False, False, False, False, False, False]])
>>> mask1 = segm.make_source_mask(size=3) >>> mask1 array([[False, False, False, False, False, False, False], [False, False, False, False, False, False, False], [False, False, True, True, True, False, False], [False, False, True, True, True, False, False], [False, False, True, True, True, False, False], [False, False, False, False, False, False, False], [False, False, False, False, False, False, False]])
>>> footprint = circular_footprint(radius=3) >>> mask2 = segm.make_source_mask(footprint=footprint) >>> mask2 array([[False, False, False, True, False, False, False], [False, True, True, True, True, True, False], [False, True, True, True, True, True, False], [ True, True, True, True, True, True, True], [False, True, True, True, True, True, False], [False, True, True, True, True, True, False], [False, False, False, True, False, False, False]])
- plot_patches(*, ax=None, origin=(0, 0), scale=1.0, labels=None, **kwargs)[source]¶
Plot the
Polygonobjects for the source segments on a matplotlibAxesinstance.- Parameters:
- ax
matplotlib.axes.AxesorNone, optional The matplotlib axes on which to plot. If
None, then the currentAxesinstance is used.- originarray_like, optional
The
(x, y)position of the origin of the displayed image.- scalefloat, optional
The scale factor applied to the polygon vertices.
- labelsint or array of int, optional
The label numbers whose polygons are to be plotted. If
None, the polygons for all labels will be plotted.- **kwargs
dict Any keyword arguments accepted by
matplotlib.patches.Polygon.
- ax
- Returns:
- patcheslist of
Polygon A list of matplotlib polygon patches for the plotted polygons. The patches can be used, for example, when adding a plot legend.
- patcheslist of
Examples
import numpy as np from photutils.segmentation import SegmentationImage data = np.array([[1, 1, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]]) segm = SegmentationImage(data) segm.imshow(figsize=(5, 5)) segm.plot_patches(edgecolor='white', lw=2)
(
Source code,png,hires.png,pdf,svg)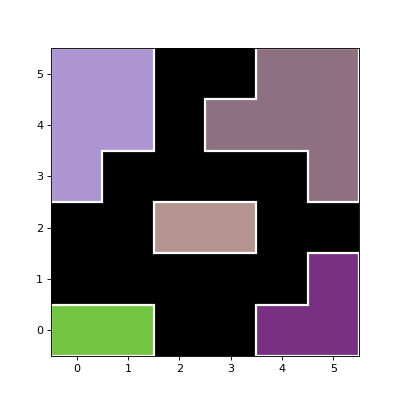
- reassign_label(label, new_label, relabel=False)[source]¶
Reassign a label number to a new number.
If
new_labelis already present in the segmentation array, then it will be combined with the inputlabelnumber. Note that this can result in a label that is no longer pixel connected.- Parameters:
- labelint
The label number to reassign.
- new_labelint
The newly assigned label number.
- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_label(label=1, new_label=2) >>> segm.data array([[2, 2, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_label(label=1, new_label=4) >>> segm.data array([[4, 4, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_label(label=1, new_label=4, relabel=True) >>> segm.data array([[2, 2, 0, 0, 2, 2], [0, 0, 0, 0, 0, 2], [0, 0, 1, 1, 0, 0], [4, 0, 0, 0, 0, 3], [4, 4, 0, 3, 3, 3], [4, 4, 0, 0, 3, 3]])
- reassign_labels(labels, new_label, relabel=False)[source]¶
Reassign one or more label numbers.
Multiple input
labelswill all be reassigned to the samenew_labelnumber. Ifnew_labelis already present in the segmentation array, then it will be combined with the inputlabels. Note that both of these can result in a label that is no longer pixel connected.- Parameters:
- labelsint, array_like (1D, int)
The label numbers(s) to reassign.
- new_labelint
The reassigned label number.
- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_labels(labels=[1, 7], new_label=2) >>> segm.data array([[2, 2, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [2, 0, 0, 0, 0, 5], [2, 2, 0, 5, 5, 5], [2, 2, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_labels(labels=[1, 7], new_label=4) >>> segm.data array([[4, 4, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [4, 0, 0, 0, 0, 5], [4, 4, 0, 5, 5, 5], [4, 4, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.reassign_labels(labels=[1, 7], new_label=2, relabel=True) >>> segm.data array([[1, 1, 0, 0, 3, 3], [0, 0, 0, 0, 0, 3], [0, 0, 2, 2, 0, 0], [1, 0, 0, 0, 0, 4], [1, 1, 0, 4, 4, 4], [1, 1, 0, 0, 4, 4]])
- relabel_consecutive(start_label=1)[source]¶
Reassign the label numbers consecutively starting from a given label number.
- Parameters:
- start_labelint, optional
The starting label number, which should be a strictly positive integer. The default is 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.relabel_consecutive() >>> segm.data array([[1, 1, 0, 0, 3, 3], [0, 0, 0, 0, 0, 3], [0, 0, 2, 2, 0, 0], [5, 0, 0, 0, 0, 4], [5, 5, 0, 4, 4, 4], [5, 5, 0, 0, 4, 4]])
- remove_border_labels(border_width, partial_overlap=True, relabel=False)[source]¶
Remove labeled segments near the array border.
Labels within the defined border region will be removed.
- Parameters:
- border_widthint
The width of the border region in pixels.
- partial_overlapbool, optional
If this is set to
True(the default), a segment that partially extends into the border region will be removed. Segments that are completely within the border region are always removed.- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_border_labels(border_width=1) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_border_labels(border_width=1, ... partial_overlap=False) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]])
- remove_label(label, relabel=False)[source]¶
Remove the label number.
The removed label is assigned a value of zero (i.e., background).
- Parameters:
- labelint
The label number to remove.
- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_label(label=5) >>> segm.data array([[1, 1, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 0], [7, 7, 0, 0, 0, 0], [7, 7, 0, 0, 0, 0]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_label(label=5, relabel=True) >>> segm.data array([[1, 1, 0, 0, 3, 3], [0, 0, 0, 0, 0, 3], [0, 0, 2, 2, 0, 0], [4, 0, 0, 0, 0, 0], [4, 4, 0, 0, 0, 0], [4, 4, 0, 0, 0, 0]])
- remove_labels(labels, relabel=False)[source]¶
Remove one or more labels.
Removed labels are assigned a value of zero (i.e., background).
- Parameters:
- labelsint, array_like (1D, int)
The label number(s) to remove.
- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_labels(labels=[5, 3]) >>> segm.data array([[1, 1, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 0, 0, 0, 0], [7, 0, 0, 0, 0, 0], [7, 7, 0, 0, 0, 0], [7, 7, 0, 0, 0, 0]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_labels(labels=[5, 3], relabel=True) >>> segm.data array([[1, 1, 0, 0, 2, 2], [0, 0, 0, 0, 0, 2], [0, 0, 0, 0, 0, 0], [3, 0, 0, 0, 0, 0], [3, 3, 0, 0, 0, 0], [3, 3, 0, 0, 0, 0]])
- remove_masked_labels(mask, partial_overlap=True, relabel=False)[source]¶
Remove labeled segments located within a masked region.
- Parameters:
- maskarray_like (bool)
A boolean mask, with the same shape as the segmentation array, where
Truevalues indicate masked pixels.- partial_overlapbool, optional
If this is set to
True(default), a segment that partially extends into a masked region will also be removed. Segments that are completely within a masked region are always removed.- relabelbool, optional
If
True, then the segmentation array will be relabeled such that the labels are in consecutive order starting from 1.
Examples
>>> from photutils.segmentation import SegmentationImage >>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> mask = np.zeros(segm.data.shape, dtype=bool) >>> mask[0, :] = True # mask the first row >>> segm.remove_masked_labels(mask) >>> segm.data array([[0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]])
>>> data = np.array([[1, 1, 0, 0, 4, 4], ... [0, 0, 0, 0, 0, 4], ... [0, 0, 3, 3, 0, 0], ... [7, 0, 0, 0, 0, 5], ... [7, 7, 0, 5, 5, 5], ... [7, 7, 0, 0, 5, 5]]) >>> segm = SegmentationImage(data) >>> segm.remove_masked_labels(mask, partial_overlap=False) >>> segm.data array([[0, 0, 0, 0, 4, 4], [0, 0, 0, 0, 0, 4], [0, 0, 3, 3, 0, 0], [7, 0, 0, 0, 0, 5], [7, 7, 0, 5, 5, 5], [7, 7, 0, 0, 5, 5]])
- reset_cmap(seed=None)[source]¶
Reset the colormap (
cmapattribute) to a new random colormap.- Parameters:
- seedint, optional
A seed to initialize the
numpy.random.BitGenerator. IfNone, then fresh, unpredictable entropy will be pulled from the OS. Separate function calls with the sameseedwill generate the same colormap.
- to_patches(*, origin=(0, 0), scale=1.0, **kwargs)[source]¶
Return a list of
Polygonobjects representing each source segment.By default, the polygon patch will have a white edge color and no face color.
- Parameters:
- originarray_like, optional
The
(x, y)position of the origin of the displayed image. This effectively translates the position of the polygons.- scalefloat, optional
The scale factor applied to the polygon vertices.
- **kwargs
dict Any keyword arguments accepted by
matplotlib.patches.Polygon.