Image Segmentation (photutils.segmentation)¶
Introduction¶
Photutils includes general-use functions to detect sources (both point-like and extended) in an image using a process called image segmentation. After detecting sources using image segmentation, we can then measure their photometry, centroids, and shape properties.
Source Extraction Using Image Segmentation¶
Image segmentation is a process of assigning a label to every pixel in an image such that pixels with the same label are part of the same source. Detected sources must have a minimum number of connected pixels that are each greater than a specified threshold value in an image. The threshold level is usually defined as some multiple of the background noise (sigma level) above the background. The image is usually filtered before thresholding to smooth the noise and maximize the detectability of objects with a shape similar to the filter kernel.
Let’s start by making a synthetic image provided by the photutils.datasets module:
>>> from photutils.datasets import make_100gaussians_image
>>> data = make_100gaussians_image()
Next, we need to subtract the background from the image. In this
example, we’ll use the Background2D class
to produce a background and background noise image:
>>> from photutils.background import Background2D, MedianBackground
>>> bkg_estimator = MedianBackground()
>>> bkg = Background2D(data, (50, 50), filter_size=(3, 3),
... bkg_estimator=bkg_estimator)
>>> data -= bkg.background # subtract the background
After subtracting the background, we need to define the detection threshold. In this example, we’ll define a 2D detection threshold image using the background RMS image. We set the threshold at the 1.5-sigma (per pixel) noise level:
>>> threshold = 1.5 * bkg.background_rms
Next, let’s convolve the data with a 2D Gaussian kernel with a FWHM of 3 pixels:
>>> from astropy.convolution import convolve
>>> from photutils.segmentation import make_2dgaussian_kernel
>>> kernel = make_2dgaussian_kernel(3.0, size=5) # FWHM = 3.0
>>> convolved_data = convolve(data, kernel)
Now we are ready to detect the sources in the background-subtracted
convolved image. Let’s find sources that have 10 connected pixels that
are each greater than the corresponding pixel-wise threshold level
defined above (i.e., 1.5 sigma per pixel above the background noise).
Note that by default “connected pixels” means “8-connected” pixels,
where pixels touch along their edges or corners. One can also use
“4-connected” pixels that touch only along their edges by setting
connectivity=4:
>>> from photutils.segmentation import detect_sources
>>> segment_map = detect_sources(convolved_data, threshold, npixels=10)
>>> print(segment_map)
<photutils.segmentation.core.SegmentationImage>
shape: (300, 500)
nlabels: 87
labels: [ 1 2 3 4 5 ... 83 84 85 86 87]
The result is a SegmentationImage
object with the same shape as the data, where detected sources are
labeled by different positive integer values. Background pixels
(non-sources) always have a value of zero. Because the segmentation
image is generated using image thresholding, the source segments
represent the isophotal footprints of each source.
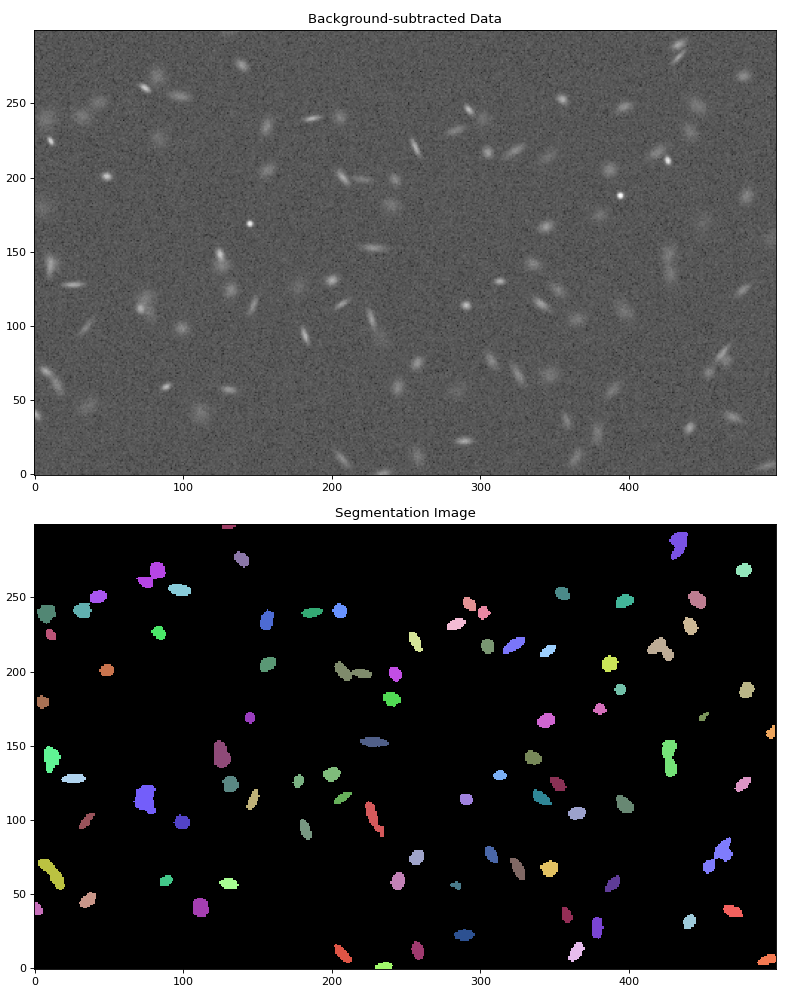
Let’s plot both the background-subtracted image and the segmentation image showing the detected sources:
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> from astropy.visualization import SqrtStretch
>>> from astropy.visualization.mpl_normalize import ImageNormalize
>>> norm = ImageNormalize(stretch=SqrtStretch())
>>> fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(10, 12.5))
>>> ax1.imshow(data, origin='lower', cmap='Greys_r', norm=norm)
>>> ax1.set_title('Background-subtracted Data')
>>> ax2.imshow(segment_map, origin='lower', cmap=segment_map.cmap,
... interpolation='nearest')
>>> ax2.set_title('Segmentation Image')
(Source code, png, hires.png, pdf, svg)
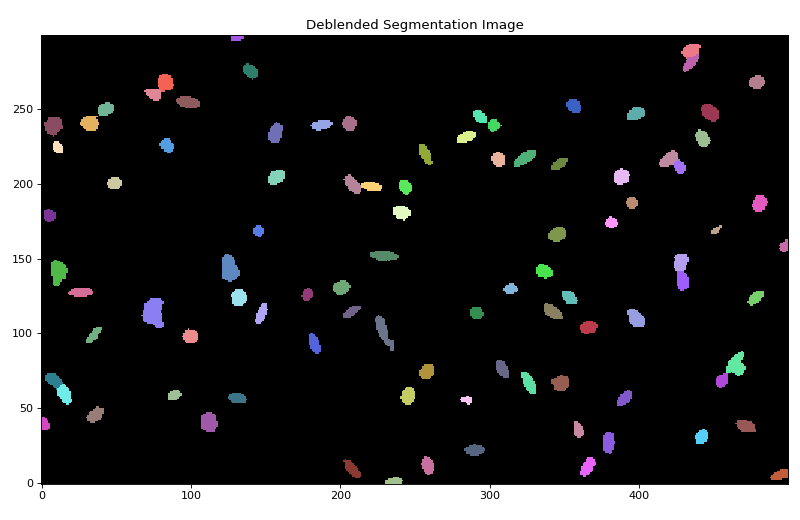
Source Deblending¶
In the example above, overlapping sources are detected as single
sources. Separating those sources requires a deblending procedure,
such as a multi-thresholding technique used by SourceExtractor.
Photutils provides a deblend_sources()
function that deblends sources uses a combination
of multi-thresholding and watershed segmentation. Note
that in order to deblend sources, they must be separated enough such
that there is a saddle point between them.
The amount of deblending can be controlled with the two
deblend_sources() keywords nlevels and
contrast. nlevels is the number of multi-thresholding levels to
use. contrast is the fraction of the total source flux that a local
peak must have to be considered as a separate object.
Here’s a simple example of source deblending:
>>> from photutils.segmentation import deblend_sources
>>> segm_deblend = deblend_sources(convolved_data, segment_map,
... npixels=10, nlevels=32, contrast=0.001,
... progress_bar=False)
where segment_map is the
SegmentationImage that was
generated by detect_sources(). Note
that the convolved_data and npixels input values should
match those used in detect_sources()
to generate segment_map. The result is a new
SegmentationImage object containing the
deblended segmentation image:
(Source code, png, hires.png, pdf, svg)
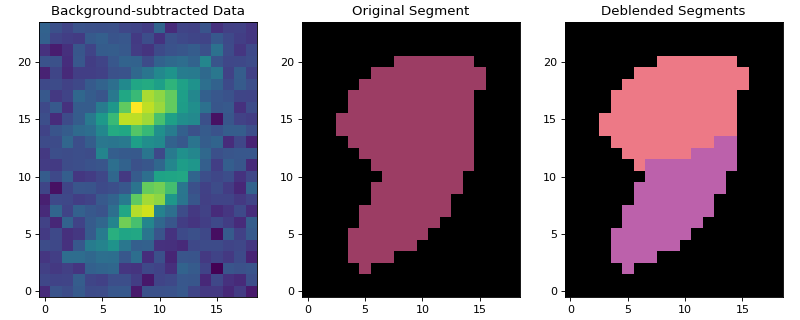
Let’s plot one of the deblended sources:
(Source code, png, hires.png, pdf, svg)
SourceFinder¶
The SourceFinder class
is a convenience class that combines the functionality
of detect_sources and
deblend_sources. After defining the object
with the desired detection and deblending parameters, you call it with
the background-subtracted (convolved) image and threshold:
>>> from photutils.segmentation import SourceFinder
>>> finder = SourceFinder(npixels=10, progress_bar=False)
>>> segment_map = finder(convolved_data, threshold)
>>> print(segment_map)
<photutils.segmentation.core.SegmentationImage>
shape: (300, 500)
nlabels: 94
labels: [ 1 2 3 4 5 ... 90 91 92 93 94]
Modifying a Segmentation Image¶
The SegmentationImage object provides
several methods that can be used to modify itself (e.g.,
combining labels, removing labels, removing border segments) prior to
measuring source photometry and other source properties, including:
reassign_label(): Reassign one or more label numbers.
relabel_consecutive(): Reassign the label numbers consecutively, such that there are no missing label numbers.
keep_labels(): Keep only the specified labels.
remove_labels(): Remove one or more labels.
remove_border_labels(): Remove labeled segments near the image border.
remove_masked_labels(): Remove labeled segments located within a masked region.
Photometry, Centroids, and Shape Properties¶
The SourceCatalog class is the primary
tool for measuring the photometry, centroids, and shape/morphological
properties of sources defined in a segmentation image. In its most
basic form, it takes as input the (background-subtracted) image and
the segmentation image. Usually the convolved image is also input,
from which the source centroids and shape/morphological properties are
measured (if not input, the unconvolved image is used instead).
Let’s continue our example from above and measure the properties of the detected sources:
>>> from photutils.segmentation import SourceCatalog
>>> cat = SourceCatalog(data, segm_deblend, convolved_data=convolved_data)
>>> print(cat)
<photutils.segmentation.catalog.SourceCatalog>
Length: 94
labels: [ 1 2 3 4 5 ... 90 91 92 93 94]
The source properties can be accessed using
SourceCatalog attributes or
output to an Astropy QTable using the
to_table() method. Please
see SourceCatalog for the many
properties that can be calculated for each source. More properties are
likely to be added in the future.
Here we’ll use the
to_table() method to
generate a QTable of source properties. Each row in the
table represents a source. The columns represent the calculated source
properties. The label column corresponds to the label value in the
input segmentation image. Note that only a small subset of the source
properties are shown below:
>>> tbl = cat.to_table()
>>> tbl['xcentroid'].info.format = '.2f' # optional format
>>> tbl['ycentroid'].info.format = '.2f'
>>> tbl['kron_flux'].info.format = '.2f'
>>> print(tbl)
label xcentroid ycentroid ... segment_fluxerr kron_flux kron_fluxerr
...
----- --------- --------- ... --------------- --------- ------------
1 235.31 1.45 ... nan 509.74 nan
2 493.92 5.79 ... nan 544.31 nan
3 207.42 9.81 ... nan 722.26 nan
4 364.86 11.11 ... nan 704.23 nan
5 258.27 11.94 ... nan 661.22 nan
... ... ... ... ... ... ...
90 419.52 216.55 ... nan 866.40 nan
91 74.55 259.86 ... nan 870.31 nan
92 82.56 267.55 ... nan 811.81 nan
93 433.88 280.75 ... nan 652.12 nan
94 434.07 288.90 ... nan 942.22 nan
Length = 94 rows
The error columns are NaN because we did not input an error array (see the Photometric Errors section below).
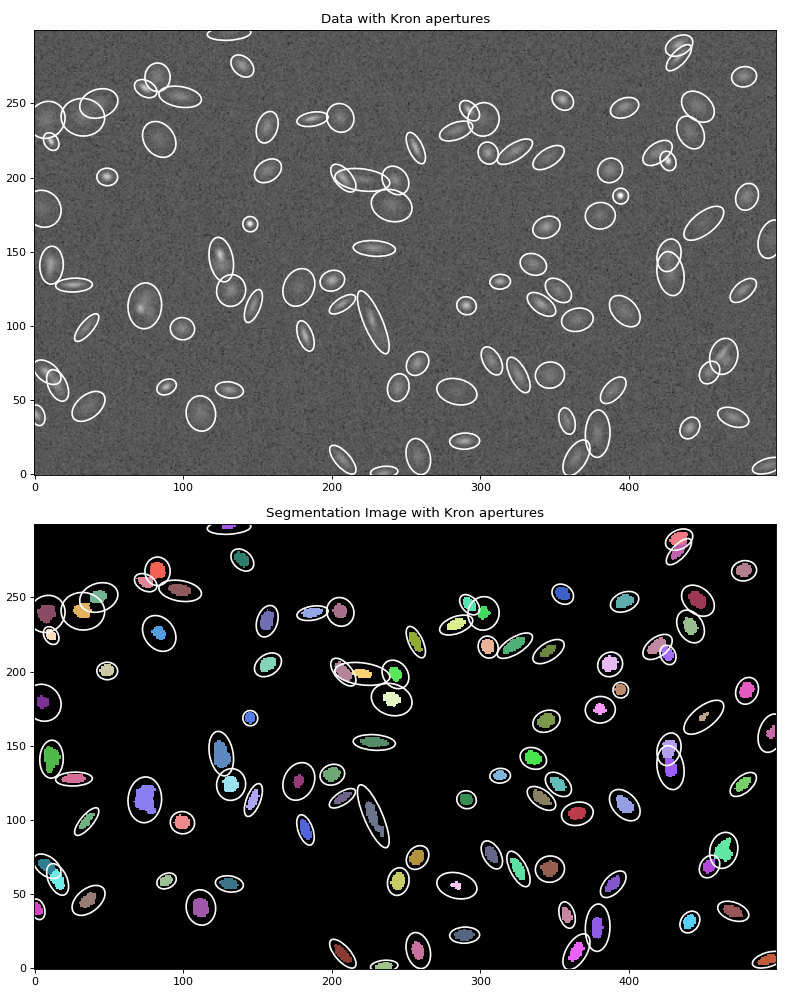
Let’s plot the calculated elliptical Kron apertures (based on the shapes of each source) on the data:
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> from astropy.visualization import simple_norm
>>> norm = simple_norm(data, 'sqrt')
>>> fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(10, 12.5))
>>> ax1.imshow(data, origin='lower', cmap='Greys_r', norm=norm)
>>> ax1.set_title('Data')
>>> ax2.imshow(segm_deblend, origin='lower', cmap=segm_deblend.cmap,
... interpolation='nearest')
>>> ax2.set_title('Segmentation Image')
>>> cat.plot_kron_apertures(ax=ax1, color='white', lw=1.5)
>>> cat.plot_kron_apertures(ax=ax2, color='white', lw=1.5)
(Source code, png, hires.png, pdf, svg)
We can also create a SourceCatalog object
containing only a specific subset of sources, defined by their
label numbers in the segmentation image:
>>> cat = SourceCatalog(data, segm_deblend, convolved_data=convolved_data)
>>> labels = [1, 5, 20, 50, 75, 80]
>>> cat_subset = cat.get_labels(labels)
>>> tbl2 = cat_subset.to_table()
>>> tbl2['xcentroid'].info.format = '.2f' # optional format
>>> tbl2['ycentroid'].info.format = '.2f'
>>> tbl2['kron_flux'].info.format = '.2f'
>>> print(tbl2)
label xcentroid ycentroid ... segment_fluxerr kron_flux kron_fluxerr
...
----- --------- --------- ... --------------- --------- ------------
1 235.31 1.45 ... nan 509.74 nan
5 258.27 11.94 ... nan 661.22 nan
20 346.99 66.83 ... nan 811.70 nan
50 5.29 178.94 ... nan 614.46 nan
75 42.96 249.88 ... nan 617.18 nan
80 130.75 297.10 ... nan 246.91 nan
By default, the to_table()
includes only a small subset of source properties. The output table
properties can be customized in the QTable using the
columns keyword:
>>> cat = SourceCatalog(data, segm_deblend, convolved_data=convolved_data)
>>> labels = [1, 5, 20, 50, 75, 80]
>>> cat_subset = cat.get_labels(labels)
>>> columns = ['label', 'xcentroid', 'ycentroid', 'area', 'segment_flux']
>>> tbl3 = cat_subset.to_table(columns=columns)
>>> tbl3['xcentroid'].info.format = '.4f' # optional format
>>> tbl3['ycentroid'].info.format = '.4f'
>>> tbl3['segment_flux'].info.format = '.4f'
>>> print(tbl3)
label xcentroid ycentroid area segment_flux
pix2
----- --------- --------- ---- ------------
1 235.3128 1.4483 47.0 445.6095
5 258.2727 11.9418 79.0 472.4109
20 346.9920 66.8323 98.0 571.3138
50 5.2910 178.9449 57.0 257.4050
75 42.9629 249.8825 79.0 428.7122
80 130.7542 297.1048 25.0 108.7877
A WCS transformation can also be input to
SourceCatalog via the wcs keyword,
in which case the sky coordinates of the source centroids can be
calculated.
Background Properties¶
Like with aperture_photometry(), the data
array that is input to SourceCatalog
should be background subtracted. If you input the background image
that was subtracted from the data into the background keyword
of SourceCatalog, the background
properties for each source will also be calculated:
>>> cat = SourceCatalog(data, segm_deblend, background=bkg.background)
>>> labels = [1, 5, 20, 50, 75, 80]
>>> cat_subset = cat.get_labels(labels)
>>> columns = ['label', 'background_centroid', 'background_mean',
... 'background_sum']
>>> tbl4 = cat_subset.to_table(columns=columns)
>>> tbl4['background_centroid'].info.format = '{:.10f}' # optional format
>>> tbl4['background_mean'].info.format = '{:.10f}'
>>> tbl4['background_sum'].info.format = '{:.10f}'
>>> print(tbl4)
label background_centroid background_mean background_sum
----- ------------------- --------------- --------------
1 5.2031964942 5.2026137494 244.5228462210
5 5.2369908637 5.2229743435 412.6149731353
20 5.2392999510 5.2702094547 516.4805265603
50 5.1910069327 5.2466148918 299.0570488310
75 5.2010736005 5.2249844756 412.7737735748
80 5.2669562370 5.2642197829 131.6054945730
Photometric Errors¶
SourceCatalog requires inputting a
total error array, i.e., the background-only error plus Poisson noise
due to individual sources. The calc_total_error()
function can be used to calculate the total error array from a
background-only error array and an effective gain.
The effective_gain, which is the ratio of counts (electrons or
photons) to the units of the data, is used to include the Poisson noise
from the sources. effective_gain can either be a scalar value or a
2D image with the same shape as the data. A 2D effective gain image
is useful for mosaic images that have variable depths (i.e., exposure
times) across the field. For example, one should use an exposure-time
map as the effective_gain for a variable depth mosaic image in
count-rate units.
Let’s assume our synthetic data is in units of electrons per
second. In that case, the effective_gain should be the
exposure time (here we set it to 500 seconds). Here we use
calc_total_error() to calculate the total error
and input it into the SourceCatalog
class. When a total error is input, the
segment_fluxerr and
kron_fluxerr properties are
calculated. segment_flux
and segment_fluxerr are the
instrumental flux and propagated flux error within the source segments:
>>> from photutils.utils import calc_total_error
>>> effective_gain = 500.0
>>> error = calc_total_error(data, bkg.background_rms, effective_gain)
>>> cat = SourceCatalog(data, segm_deblend, error=error)
>>> labels = [1, 5, 20, 50, 75, 80]
>>> cat_subset = cat.get_labels(labels) # select a subset of objects
>>> columns = ['label', 'xcentroid', 'ycentroid', 'segment_flux',
... 'segment_fluxerr']
>>> tbl5 = cat_subset.to_table(columns=columns)
>>> tbl5['xcentroid'].info.format = '{:.4f}' # optional format
>>> tbl5['ycentroid'].info.format = '{:.4f}'
>>> tbl5['segment_flux'].info.format = '{:.4f}'
>>> tbl5['segment_fluxerr'].info.format = '{:.4f}'
>>> for col in tbl5.colnames:
... tbl5[col].info.format = '%.8g' # for consistent table output
>>> print(tbl5)
label xcentroid ycentroid segment_flux segment_fluxerr
----- --------- --------- ------------ ---------------
1 235.24243 1.2020749 445.60947 14.634553
5 258.29728 11.899213 472.41087 19.060458
20 346.94871 66.802889 571.31384 21.674279
50 5.2694265 178.9268 257.40504 16.431396
75 43.030177 249.92847 428.71216 19.175229
80 130.64209 297.11218 108.78768 10.882015
Pixel Masking¶
Pixels can be completely ignored/excluded (e.g., bad pixels) when
measuring the source properties by providing a boolean mask image
via the mask keyword (True pixel values are masked) to the
SourceCatalog class. Note that
non-finite data values (NaN and inf) are automatically masked.
Filtering¶
SourceExtractor’s centroid and morphological parameters are
always calculated from a convolved, or filtered, “detection” image
(convolved_data), i.e., the image used to define the segmentation
image. The usual downside of the filtering is the sources will be
made more circular than they actually are. If you wish to reproduce
SourceExtractor centroid and morphology results, then input the
convolved_data (or kernel, but not both). If convolved_data
and kernel are both None, then the unfiltered data will be
used for the source centroid and morphological parameters. Note that
photometry is always performed on the unfiltered data.
Reference/API¶
This subpackage contains tools for detecting sources using image segmentation and measuring their centroids, photometry, and morphological properties.
Functions¶
|
Deblend overlapping sources labeled in a segmentation image. |
|
Detect sources above a specified threshold value in an image. |
|
Calculate a pixel-wise threshold image that can be used to detect sources. |
|
Make a normalized 2D circular Gaussian kernel. |
Classes¶
|
Class for a single labeled region (segment) within a segmentation image. |
|
Class for a segmentation image. |
|
Class to create a catalog of photometry and morphological properties for sources defined by a segmentation image. |
|
Class to detect sources, including deblending, in an image using segmentation. |