ImageDepth¶
- class photutils.utils.ImageDepth(aper_radius, *, nsigma=5.0, mask_pad=0, napers=1000, niters=10, overlap=True, overlap_maxiters=100, seed=None, zeropoint=0.0, sigma_clip=SigmaClip(sigma=3.0, sigma_lower=3.0, sigma_upper=3.0, maxiters=10, cenfunc='median', stdfunc='std', grow=False), progress_bar=True)[source]¶
Bases:
objectClass to calculate the limiting flux and magnitude of an image.
- Parameters:
- aper_radiusfloat
The radius (in pixels) of the circular apertures used to compute the image depth.
- nsigmafloat, optional
The number of standard deviations at which to compute the image depths.
- mask_padfloat, optional
An additional padding (in pixels) to apply when dilating the input mask.
- napersint, optional
The number of circular apertures used to compute the image depth.
- nitersint, optional
The number of iterations, each with randomly-generated apertures, for which the image depth will be calculated.
- overlapbool, optional
Whether to allow the apertures to overlap.
- overlap_maxitersint, optional
The maximum number of iterations that will be used when attempting to find additional non-overlapping apertures. This keyword has no effect unless
overlap=False. While increasing this number may generate more non-overlapping apertures in crowded cases, it will also run slower.- seedint, optional
A seed to initialize the
numpy.random.BitGenerator. IfNone, then fresh, unpredictable entropy will be pulled from the OS. Separate function calls with the sameseedwill generate the same results.- zeropointfloat, optional
The zeropoint used to calculate the magnitude limit from the flux limit:
\[m_{\mathrm{lim}} = -2.5 \log_{10} f_{\mathrm{lim}} + \mathrm{zeropoint}\]- sigma_clip
astropy.stats.SigmaClipinstance, optional A
SigmaClipobject that defines the sigma clipping parameters to use when computing the limiting flux. IfNonethen no sigma clipping will be performed.- progress_barbool, optional
Whether to display a progress bar. The progress bar requires that the tqdm optional dependency be installed. Note that the progress bar does not currently work in the Jupyter console due to limitations in
tqdm.
Notes
The image depth is calculated by placing random circular apertures with the specified radius on blank regions of the image. The number of apertures is specified by the
naperskeyword. The blank regions are calculated from an input mask, which should mask both sources in the image and areas without image coverage. The input mask will be dilated with a circular footprint with a radius equal to the inputaper_radiusplusmask_pad. The image border is also masked with the same radius.The flux limit is calculated as the standard deviation of the aperture fluxes times the input
nsigmasignificance level. The aperture flux values can be sigma clipped prior to computing the standard deviation using thesigma_clipkeyword.The flux limit is calculated
niterstimes, each with a randomly-generated set of circular apertures. The returned flux limit is the average of these flux limits.The magnitude limit is calculated from flux limit using the input
zeropointkeyword as:\[m_{\mathrm{lim}} = -2.5 \log_{10} f_{\mathrm{lim}} + \mathrm{zeropoint}\]Examples
>>> from astropy.convolution import convolve >>> from astropy.visualization import simple_norm >>> from photutils.datasets import make_100gaussians_image >>> from photutils.segmentation import SourceFinder, make_2dgaussian_kernel >>> from photutils.utils import ImageDepth >>> bkg = 5.0 >>> data = make_100gaussians_image() - bkg >>> kernel = make_2dgaussian_kernel(3.0, size=5) >>> convolved_data = convolve(data, kernel) >>> npixels = 10 >>> threshold = 3.2 >>> finder = SourceFinder(npixels=npixels, progress_bar=False) >>> segment_map = finder(convolved_data, threshold) >>> mask = segment_map.make_source_mask() >>> radius = 4 >>> depth = ImageDepth(radius, nsigma=5.0, napers=500, niters=2, ... mask_pad=5, overlap=False, seed=123, ... zeropoint=23.9, progress_bar=False) >>> limits = depth(data, mask) >>> print(limits) (68.0112578151062, 19.318547982855563)
# plot the random apertures for the first iteration import matplotlib.pyplot as plt from astropy.convolution import convolve from astropy.visualization import simple_norm from photutils.datasets import make_100gaussians_image from photutils.segmentation import SourceFinder, make_2dgaussian_kernel from photutils.utils import ImageDepth bkg = 5.0 data = make_100gaussians_image() - bkg kernel = make_2dgaussian_kernel(3.0, size=5) convolved_data = convolve(data, kernel) npixels = 10 threshold = 3.2 finder = SourceFinder(npixels=npixels, progress_bar=False) segment_map = finder(convolved_data, threshold) mask = segment_map.make_source_mask() radius = 4 depth = ImageDepth(radius, nsigma=5.0, napers=500, niters=2, overlap=False, seed=123, progress_bar=False) limits = depth(data, mask) fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(9, 3)) norm = simple_norm(data, 'sqrt', percent=99.) ax[0].imshow(data, norm=norm) color = 'orange' depth.apertures[0].plot(ax[0], color=color) ax[0].set_title('Data with blank apertures') ax[1].imshow(mask, interpolation='none') depth.apertures[0].plot(ax[1], color=color) ax[1].set_title('Mask with blank apertures') plt.subplots_adjust(left=0.05, right=0.98, bottom=0.05, top=0.95, wspace=0.15)
(
Source code,png,hires.png,pdf,svg)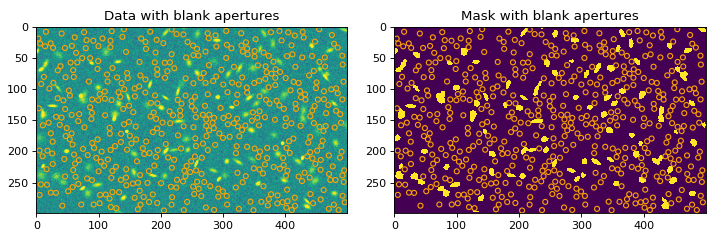
- Attributes:
- apertureslist of
CircularAperture A list of circular apertures for each iteration.
- napers_usedint
A list of the number of apertures used for each iteration.
- fluxeslist of
ndarray A list of arrays containing the flux measurements for each iteration.
- flux_limits1D
ndarray An array of the flux limits for each iteration.
- mag_limits1D
ndarray An array of the magnitude limits for each iteration.
- apertureslist of
Methods Summary
__call__(data, mask)Calculate the limiting flux and magnitude of an image.
Methods Documentation
- __call__(data, mask)[source]¶
Calculate the limiting flux and magnitude of an image.
- Parameters:
- data2D
ndarray The 2D array, which should be in flux units (not surface brightness units).
- mask2D bool
ndarray A 2D mask array with the same shape as
datawhere aTruevalue indicates the corresponding element ofdatais masked. The input array should mask both sources (e.g., from a segmentation image) and regions without image coverage. IfNone, then the entire image will be used.
- data2D
- Returns:
- flux_limit, mag_limitfloat
The flux and magnitude limits. The flux limit is returned in the same units as the input
data. The magnitude limit is calculated from the flux limit and the inputzeropoint.